We structure RegenBase data using the RegenBase ontology, BioAssay Ontology, Units of Measurement Ontology, and Ontology for Biomedical Investigations. To illustrate how we use these ontologies to model events, neurite outgrowth assay data, kinase inhibition assay data, and gene expression assay data, we show examples of each below with a detailed description.
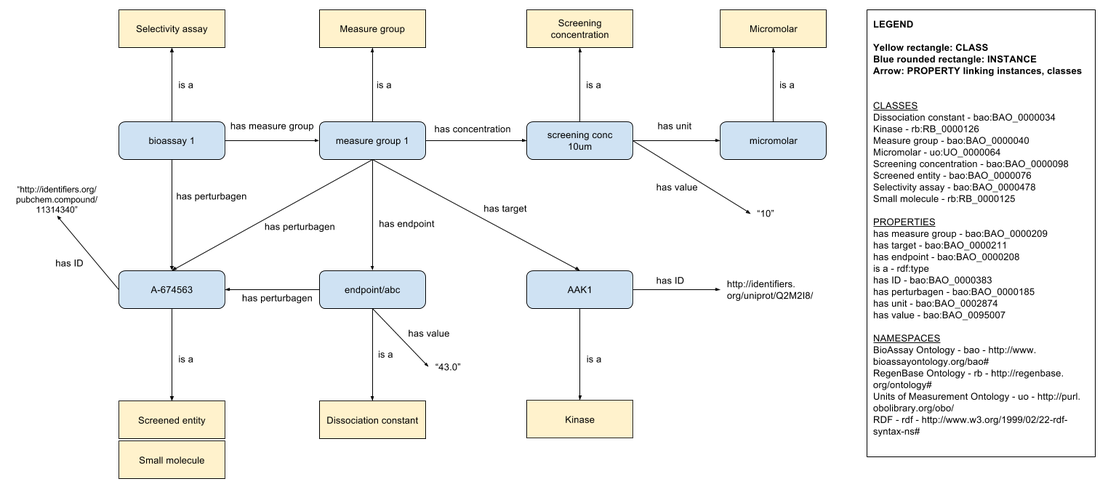
Kinase inhibition assay data
For each kinase inhibition assay data point, we create:
The 'selectivity assay' individual is linked to the 'measure group' individual with the 'has measure group' property. The 'measure group' is linked to the measured 'dissociation constant' endpoint with the 'has endpoint' property. The 'selectivity assay', 'measure group', and 'dissociation constant' individuals are each linked to a 'perturbagen' individual with the 'has perturbagen' property. The 'measure group' is also linked to the 'screening concentration' individual with the 'has 'concentration' property. The 'screening concentration' individual is linked to its unit and value. Finally, the 'measure group' is linked to the 'Kinase' target of the assay with the 'has target' property. Both perturbagen and kinase are linked to a resolvable identifier with the 'has ID' property.
Kinase inhibition assay data
For each kinase inhibition assay data point, we create:
- an instance of the BioAssay Ontology 'Selectivity assay' class to represent the assay carried out
- a 'Measure group' instance which is used to link the assay to its results
- a 'Dissociation constant' instance to represent the assay result
- a 'Screened entity'/'Small molecule' instance to represent the kinase inhibitor, and
- a 'Kinase' instance to represent the kinase whose activity is measured by the assay
- a 'Screening concentration' instance to quantify the concentration of the kinase inhibitor used in the assay
The 'selectivity assay' individual is linked to the 'measure group' individual with the 'has measure group' property. The 'measure group' is linked to the measured 'dissociation constant' endpoint with the 'has endpoint' property. The 'selectivity assay', 'measure group', and 'dissociation constant' individuals are each linked to a 'perturbagen' individual with the 'has perturbagen' property. The 'measure group' is also linked to the 'screening concentration' individual with the 'has 'concentration' property. The 'screening concentration' individual is linked to its unit and value. Finally, the 'measure group' is linked to the 'Kinase' target of the assay with the 'has target' property. Both perturbagen and kinase are linked to a resolvable identifier with the 'has ID' property.
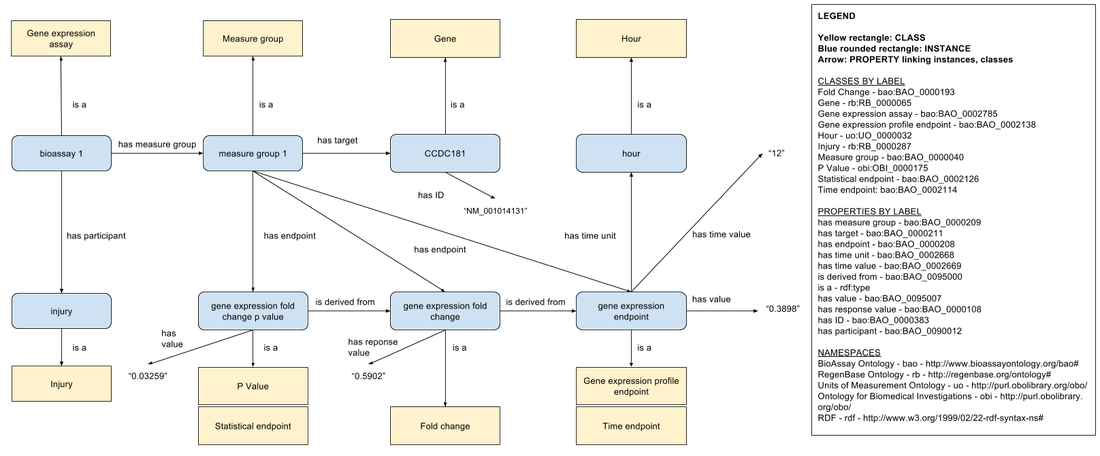
Gene expression assay data
For each gene expression assay data point, we create:
For each gene expression assay data point, we create:
- an instance of the BioAssay Ontology 'Gene expression assay' class
- a 'Measure group' instance to link the assay to its results
- an 'Injury' instance to indicate that the gene expression is being measured after an injury
- a 'Gene' instance to represent the gene whose expression is measured by the assay
- a 'Gene expression profile endpoint'/'Time endpoint' instance to represent the gene expression measured after injury
- a 'Gene expression profile endpoint'/'Time endpoint' instance to represent the control gene expression value (not shown for brevity, but included in the data)
- an 'Hour' instance to represent the time point after injury at which the gene expression values were measured
- a 'Fold change' instance to represent the fold change value derived from comparing the injury-induced gene expression value to a control gene expression value
- a 'P Value'/'Statistical endpoint' instance to represent the statistical significance of the fold change