The RegenBase Ontology: Formalizing SCI biology
Ontologies define concepts and the relationships between them in a way that allows computers to ask questions and make inferences. A well constructed ontology allows researchers to leverage computational power for categorizing and querying large amounts of data.
We have developed the RegenBase Ontology to describe the entities (classes) in the domain of SCI and the relationships between them. A short paper outlining the RegenBase Ontology was published in Neural Regeneration Research (2014).
The Regenbase Ontology was published online in the National Center for Biomedical Ontology BioPortal in 2014. It uses Description Logics (DL) with SROIQ(D) expressivity.
Expressivity refers to the types of relationships that can exist in an ontology. The most widely used ontology in biology, the Gene Ontology, has limited relationships. These include is a (is a subtype of); part of; has part; regulates, negatively regulates and positively regulates. The RegenBase ontology has additional relationships with more complex logic. It is our hope that this will allow users to ask more complex questions about SCI outcomes and axon regeneration.
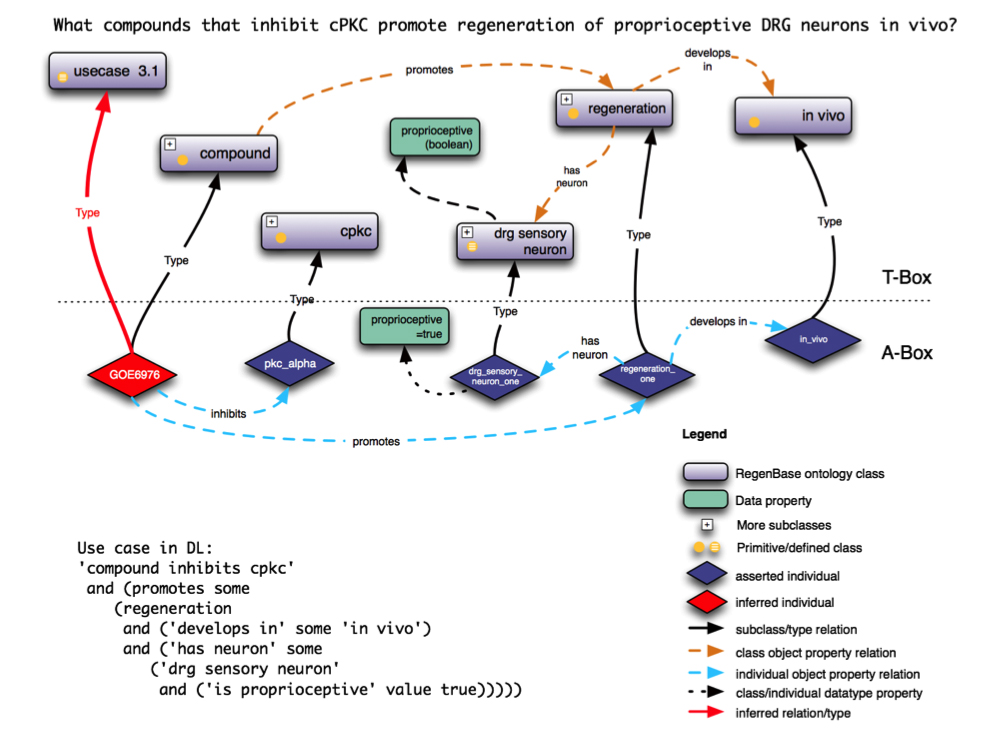
The illustration below shows an example DL expression for describing entities that inhibit a specific enzyme (CPKC) and promote regeneration of proprioceptive DRG sensory neurons. These kind of expressions, as well as queries in SPARQL, can be used to query the RegenBase knowledge base through ontologies.
We have developed the RegenBase Ontology to describe the entities (classes) in the domain of SCI and the relationships between them. A short paper outlining the RegenBase Ontology was published in Neural Regeneration Research (2014).
The Regenbase Ontology was published online in the National Center for Biomedical Ontology BioPortal in 2014. It uses Description Logics (DL) with SROIQ(D) expressivity.
Expressivity refers to the types of relationships that can exist in an ontology. The most widely used ontology in biology, the Gene Ontology, has limited relationships. These include is a (is a subtype of); part of; has part; regulates, negatively regulates and positively regulates. The RegenBase ontology has additional relationships with more complex logic. It is our hope that this will allow users to ask more complex questions about SCI outcomes and axon regeneration.
The illustration below shows an example DL expression for describing entities that inhibit a specific enzyme (CPKC) and promote regeneration of proprioceptive DRG sensory neurons. These kind of expressions, as well as queries in SPARQL, can be used to query the RegenBase knowledge base through ontologies.